You are here: Foswiki>DataServices Web>SkimTest061212 (08 Jan 2007, MarcoMambelli)Edit Attach
SkimTest061212
Description
This tests executes the chain done on a smaller sample in DssPrototype. Here the skim is executed on the whole dataset (instead of 250 elements - 20 positive samples) and using submitted jobs (Condor or Grid). The execution and validation are not fully automatic even if these are more automatic than the one in DssPrototype. To concatenate inputs and plot them in root:
TChain* tags = new TChain();
tags->AddFile("/local/workdir/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968._01-04.root",TChain::kBigNumber,"CollectionTree");
tags->AddFile("/local/workdir/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968._05-09.root",TChain::kBigNumber,"CollectionTree");
tags->AddFile("/local/workdir/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968._10-13.root",TChain::kBigNumber,"CollectionTree");
tags->AddFile("/local/workdir/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968._14-17.root",TChain::kBigNumber,"CollectionTree");
tags->AddFile("/local/workdir/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968._18-21.root",TChain::kBigNumber,"CollectionTree");
tags->AddFile("/local/workdir/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968._22-25.root",TChain::kBigNumber,"CollectionTree");
tags->AddFile("/local/workdir/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968._26-30.root",TChain::kBigNumber,"CollectionTree");
tags->AddFile("/local/workdir/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968._31-34.root",TChain::kBigNumber,"CollectionTree");
tags->AddFile("/local/workdir/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968._35-38.root",TChain::kBigNumber,"CollectionTree");
tags->AddFile("/local/workdir/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968/testIdeal_07.005711.PythiaB_bbmu6mu4X.recon.TAG.v12000201_tid002968._39-50.root",TChain::kBigNumber,"CollectionTree");
tags.Print()
tags.Draw("NJet")
tags.Draw("NLooseElectron")
tags.Draw("NJet", "NJet>0&&NLooseElectron>0")
tags.Draw("NJet", "NLooseElectron>0")
tags.Draw("NLooseElectron", "NJet>0&&NLooseElectron>0")
tags.Draw("NLooseElectron", "NJet>0")
Results are produced using condorjobsubmit.sh or gridjobsubmit.sh that run dsspilot.sh (attached: dsspilot.sh) on the worker nodes.
The script submitloop.py is a simple loop to submit multiple jobs.
To concatenate the results and plot them (in root):
TChain* outtags = new TChain();
outtags->AddFile("/share/data/t2data/results2/mytest_0_400.TAG.root",TChain::kBigNumber,"CollectionTree");
outtags->AddFile("/share/data/t2data/results2/mytest_400_400.TAG.root",TChain::kBigNumber,"CollectionTree");
outtags->AddFile("/share/data/t2data/results2/mytest_800_400.TAG.root",TChain::kBigNumber,"CollectionTree");
outtags->AddFile("/share/data/t2data/results2/mytest_1200_400.TAG.root",TChain::kBigNumber,"CollectionTree");
outtags->AddFile("/share/data/t2data/results2/mytest_1600_400.TAG.root",TChain::kBigNumber,"CollectionTree");
outtags->AddFile("/share/data/t2data/results2/mytest_2000_400.TAG.root",TChain::kBigNumber,"CollectionTree");
outtags->AddFile("/share/data/t2data/results2/mytest_2400_400.TAG.root",TChain::kBigNumber,"CollectionTree");
outtags->AddFile("/share/data/t2data/results2/mytest_2800_400.TAG.root",TChain::kBigNumber,"CollectionTree");
outtags->AddFile("/share/data/t2data/results2/mytest_3200_400.TAG.root",TChain::kBigNumber,"CollectionTree");
outtags->AddFile("/share/data/t2data/results2/mytest_3600_400.TAG.root",TChain::kBigNumber,"CollectionTree");
outtags->AddFile("/share/data/t2data/results2/mytest_4000_400.TAG.root",TChain::kBigNumber,"CollectionTree");
outtags.Print()
outtags.Draw("NJet")
outtags.Draw("NLooseElectron")
To compare the input data subject to the cut to the output data you can use the attached root macro in doplot.C.
-- MarcoMambelli - 13 Dec 2006
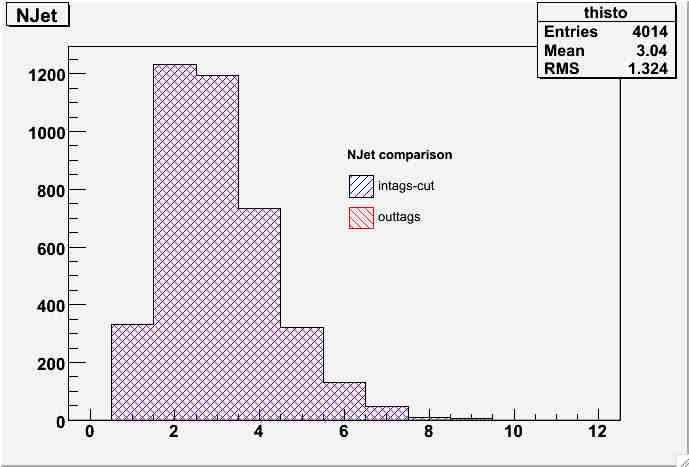
- NJet comparison:
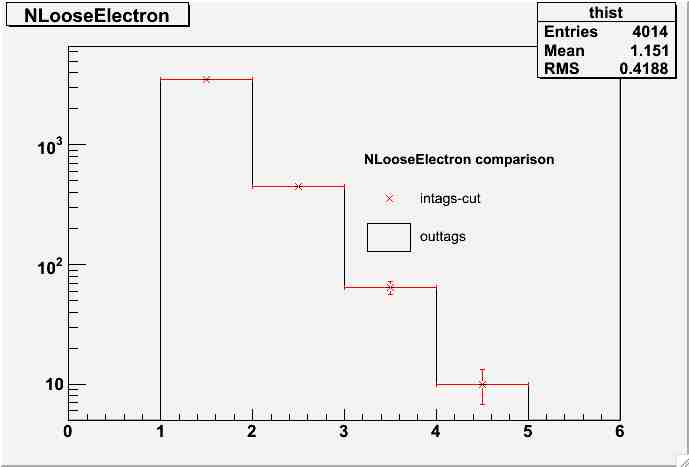
- NLoosElectron comparison:
| I | Attachment | Action | Size | Date | Who | Comment |
|---|---|---|---|---|---|---|
| |
condorjobsubmit.sh | manage | 1002 bytes | 19 Dec 2006 - 18:27 | MarcoMambelli | condor submission |
| |
doplot.C | manage | 4 K | 19 Dec 2006 - 18:20 | MarcoMambelli | result plots and comparison |
| |
dsspilot.sh | manage | 2 K | 14 Dec 2006 - 21:43 | MarcoMambelli | pilot to execute ds job |
| |
gridjobsubmit.sh | manage | 1 K | 19 Dec 2006 - 18:28 | MarcoMambelli | grid submission |
| |
njet-comp.jpg | manage | 30 K | 08 Jan 2007 - 17:09 | MarcoMambelli | NJet comparison |
| |
nloose-comp.jpg | manage | 17 K | 08 Jan 2007 - 17:10 | MarcoMambelli | NLoosElectron comparison |
Edit | Attach | Print version | History: r5 < r4 < r3 < r2 | Backlinks | View wiki text | Edit wiki text | More topic actions
Topic revision: r5 - 08 Jan 2007, MarcoMambelli
 Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors.
Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors. Ideas, requests, problems regarding Foswiki? Send feedback


